- 2012-04-22 (日) 18:39
- DBCLS
Search for amino acids included in Figures
Not only nucleotide sequences, but also amino acid sequences appearing in article figures can be quickly searched using GGRNA.
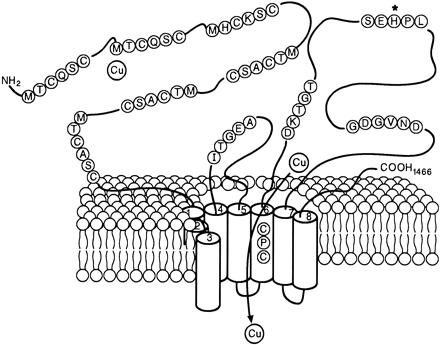
[Schaefer et al. (1999) IV. Wilson’s disease and Menkes disease. Am. J. Physiol. Gastrointest. Liver Physiol. 276, G311-G314]
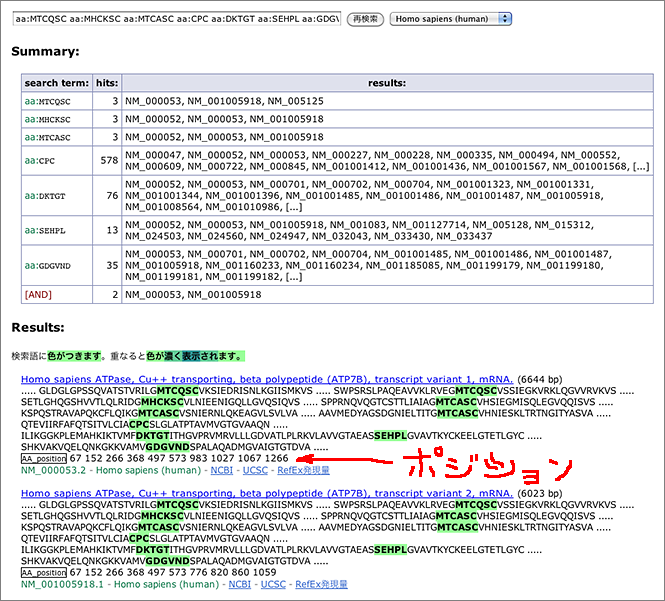
To search for the protein illustrated in the above figure, just enter partial amino acid sequence to the search box like [ aa:MTCQSC ] (→ GGRNA) or [ aa:MHCKSC ] (→ GGRNA) and you can get results. GGRNA works case insensitively, so [ AA:mhcksc ] will do the same.
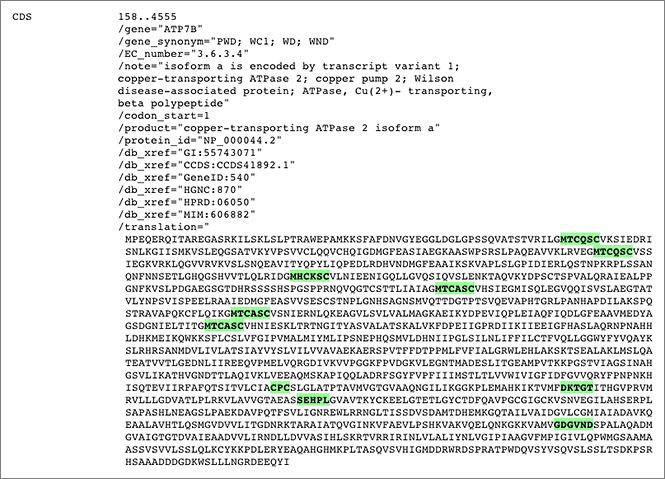
GGRNA works great for not only searching genes, but also for locating the sequences. You can enter all the sequences shown in the figure like [ aa:MTCQSC aa:MHCKSC aa:MTCASC aa:CPC aa:DKTGT aa:SEHPL aa:GDGVND ] (→ GGRNA) to easily see where they are located.
Search caspase cleavage sites
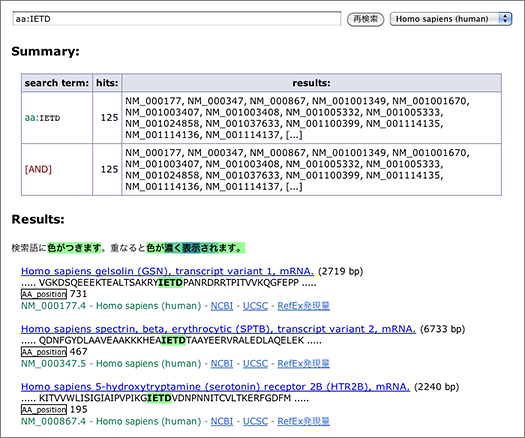
Caspases are a group of cystein proteases involved in the induction of apoptosis, and they recognize certain peptide sequences to cleave proteins. By using GGRNA amino-acid search, you can find genes whose products are likely to be cleaved by capspases, and locate their cleavage sites. Shown below are the cleavage sites of caspase-3 and caspase-8. Note that GGRNA works in case insensitive manner, so [ aa:DEVD ] and [ AA:devd ] will return the same results.
In the amino-acid sequence displayed, you can see IETD with green highlight. Below that, the position corresponding to the query sequence is indicated as ‘AA_position 731.’ Please be aware that the number indicates the position of the first matched residue (in this case I), not the residue that gets cleaved (D).
Search for ER localization signals
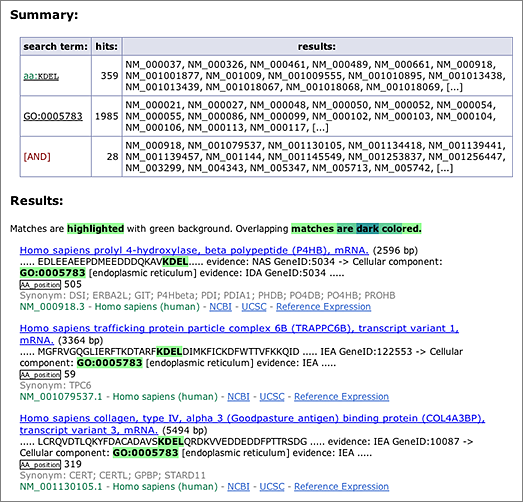
The amino acid sequence ‘KDEL’ at the C-terminus serves as the endoplasmic reticulum (ER) retention signal. To search for the KDEL motif in GGRNA, enter [ aa:KDEL ] in the search box (→ GGRNA). An operator aa: restricts the search to within amino acid sequences only. Searching [ aa:KDEL ] in human will retrieve 359 results (as of RefSeq release 52, Mar. 2012), but these results contain KDELs that are not at the C-terminus.
On the other hand, transcripts annotated as GO:0005783 (endoplasmic reticulum) can be retrieved by searching [ GO:0005783 ] (→ GGRNA), which returns 1,985 results.
An intersection of these two searches can be obtained by entering the two keywords separated by a space: [ aa:KDEL GO:0005783 ] (→ GGRNA), which returns 28 results. Of these, 13 results contain the KDEL motif at the C-terminus. Searching by sequences and other keywords simultaneously is one of the unique advantages of GGRNA.
Search for plant peptide hormones
Search for plant peptide hormones using signal sequences (by @hkanekane).
- CLV3 → [ aa:PDPLHHH ] (→ Search for Arabidopsis thaliana genes using GGRNA)
- RGF → [ aa:PPRHN ] (→ Search for Arabidopsis thaliana genes using GGRNA)
For sequences this short, BLAST does not work very well. GGRNA does overwhelmingly better than BLAST in searching short sequences consisting of less than 5 amino acids, except that GGRNA cannot do fuzzy search. I’m planning to implement GGRNA with amino-acid fuzzy search near future.
- Newer: GGRNAの論文掲載+今後の計画
- Older: GGRNA Search Examples (DNA sequences)
Trackbacks:0
- Trackback URL for this entry
- /archives/1654/trackback
- Listed below are links to weblogs that reference
- GGRNA Search Examples (protein sequences) from mesoの実験ノート