Home > Archives > 2012-04
2012-04
GGRNA Search Examples (protein sequences)
- 2012-04-22 (日)
- DBCLS
Search for amino acids included in Figures
Not only nucleotide sequences, but also amino acid sequences appearing in article figures can be quickly searched using GGRNA.
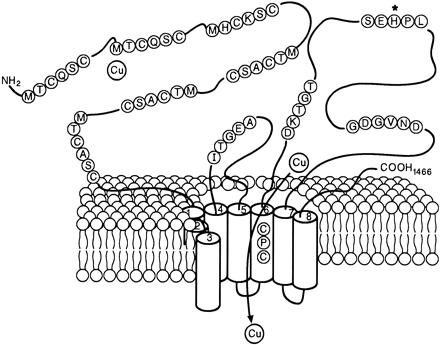
[Schaefer et al. (1999) IV. Wilson’s disease and Menkes disease. Am. J. Physiol. Gastrointest. Liver Physiol. 276, G311-G314]
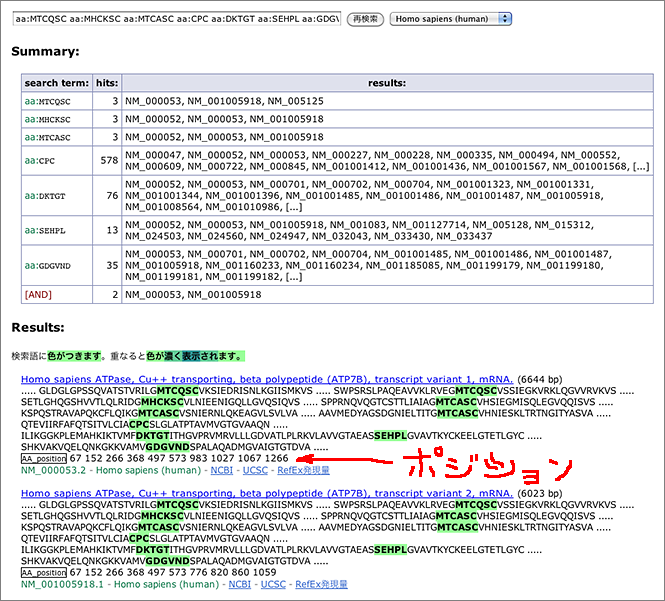
To search for the protein illustrated in the above figure, just enter partial amino acid sequence to the search box like [ aa:MTCQSC ] (→ GGRNA) or [ aa:MHCKSC ] (→ GGRNA) and you can get results. GGRNA works case insensitively, so [ AA:mhcksc ] will do the same.
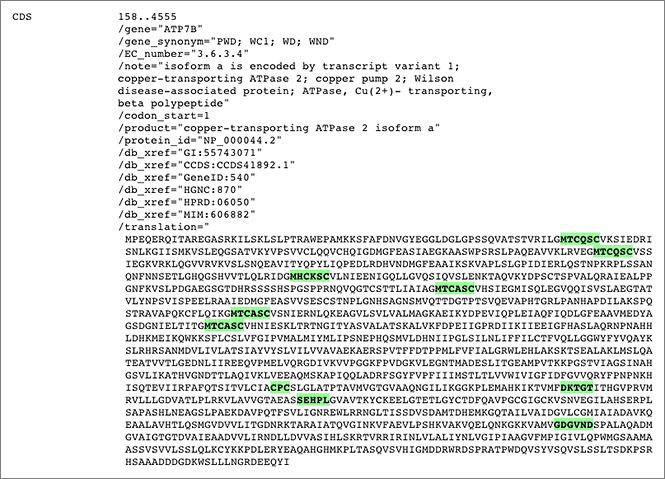
GGRNA works great for not only searching genes, but also for locating the sequences. You can enter all the sequences shown in the figure like [ aa:MTCQSC aa:MHCKSC aa:MTCASC aa:CPC aa:DKTGT aa:SEHPL aa:GDGVND ] (→ GGRNA) to easily see where they are located.
Search caspase cleavage sites
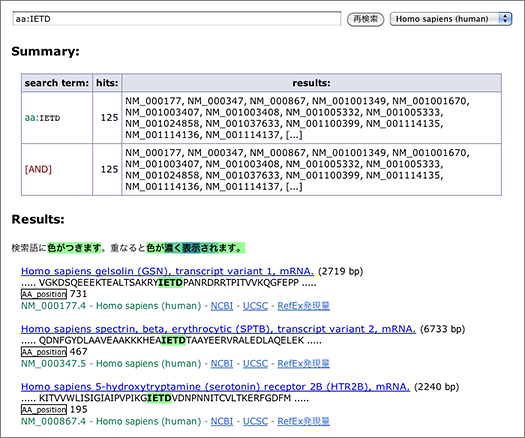
Caspases are a group of cystein proteases involved in the induction of apoptosis, and they recognize certain peptide sequences to cleave proteins. By using GGRNA amino-acid search, you can find genes whose products are likely to be cleaved by capspases, and locate their cleavage sites. Shown below are the cleavage sites of caspase-3 and caspase-8. Note that GGRNA works in case insensitive manner, so [ aa:DEVD ] and [ AA:devd ] will return the same results.
In the amino-acid sequence displayed, you can see IETD with green highlight. Below that, the position corresponding to the query sequence is indicated as ‘AA_position 731.’ Please be aware that the number indicates the position of the first matched residue (in this case I), not the residue that gets cleaved (D).
Search for ER localization signals
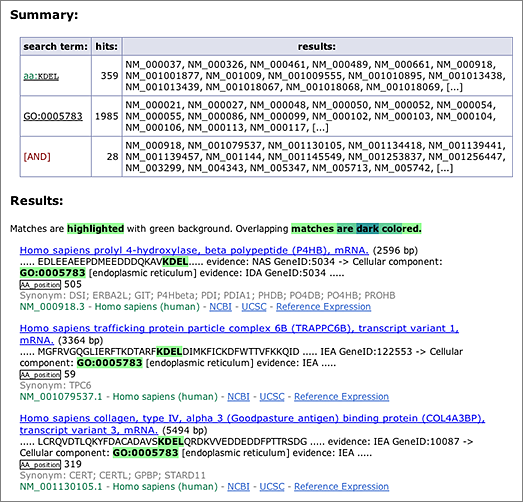
The amino acid sequence ‘KDEL’ at the C-terminus serves as the endoplasmic reticulum (ER) retention signal. To search for the KDEL motif in GGRNA, enter [ aa:KDEL ] in the search box (→ GGRNA). An operator aa: restricts the search to within amino acid sequences only. Searching [ aa:KDEL ] in human will retrieve 359 results (as of RefSeq release 52, Mar. 2012), but these results contain KDELs that are not at the C-terminus.
On the other hand, transcripts annotated as GO:0005783 (endoplasmic reticulum) can be retrieved by searching [ GO:0005783 ] (→ GGRNA), which returns 1,985 results.
An intersection of these two searches can be obtained by entering the two keywords separated by a space: [ aa:KDEL GO:0005783 ] (→ GGRNA), which returns 28 results. Of these, 13 results contain the KDEL motif at the C-terminus. Searching by sequences and other keywords simultaneously is one of the unique advantages of GGRNA.
Search for plant peptide hormones
Search for plant peptide hormones using signal sequences (by @hkanekane).
- CLV3 → [ aa:PDPLHHH ] (→ Search for Arabidopsis thaliana genes using GGRNA)
- RGF → [ aa:PPRHN ] (→ Search for Arabidopsis thaliana genes using GGRNA)
For sequences this short, BLAST does not work very well. GGRNA does overwhelmingly better than BLAST in searching short sequences consisting of less than 5 amino acids, except that GGRNA cannot do fuzzy search. I’m planning to implement GGRNA with amino-acid fuzzy search near future.
- Comments (Close): 0
- Trackbacks: 0
GGRNA Search Examples (DNA sequences)
- 2012-04-22 (日)
- DBCLS
One of the strength of GGRNA is the ultrafast search of nucleotide and amino-acid sequences. Let’s learn how to search them.
In silico PCR: search for PCR primer binding sites
Have you ever thought “I just want to use the exact primers in this paper for PCR,” or “Oops I can’t remember which region my primers amplify?” Here’s how GGRNA can help.
Let’s say you want to use the following PCR primers:
Forward primer: CTAGCTGCCAAAGAAGGACAT
Reverse primer: CAATGAGATGTTGTCGTGCTC
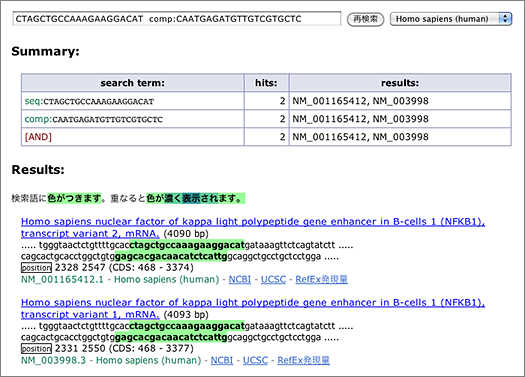
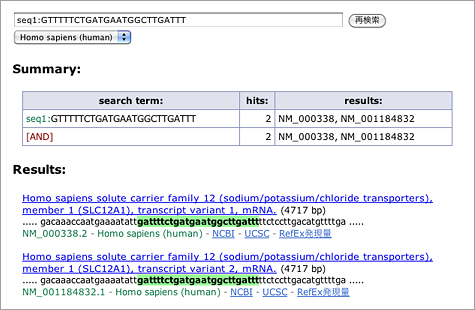
Enter [ CTAGCTGCCAAAGAAGGACAT comp:CAATGAGATGTTGTCGTGCTC ] to the search box (→ GGRNA). Since reverse primer is designed against complementary sequence, add comp: operator to let GGRNA to search for complementary sequences.
In this case, two transcript variants of NFκB (NM_001165412.1, NM_003998.3) are retrieved. Let’s take a look at the first one (NM_001165412.1).
Look at the nucleotides highlighted with green: below the highlighted letters, you can see “position 2328 2547,” indicating the start positions of the sequences matched with the queried primers. You can calculate the size of the amplified product by using these numbers: 2547 – 2328 + 21 = 240 (bp), where 21 is the length of the reverse primer. You can also see (CDS: 468 – 3374) at the right side of “position,” which indicates the corresponding CDS region. It means that the primers used here are designed within CDS.
Now try to find out the length of the second (NM_003998.3) amplified products — yes, it’s 2550 – 2331 + 21 = 240 (bp)! This NFκB transcript variant seems to give products with the same size.
Let’s have a close look at the primer binding sites. Click the title of the first hit, “Homo sapiens nuclear factor of kappa …,” and the details of the amplified product are displayed, including the primer binding sites highlighted.
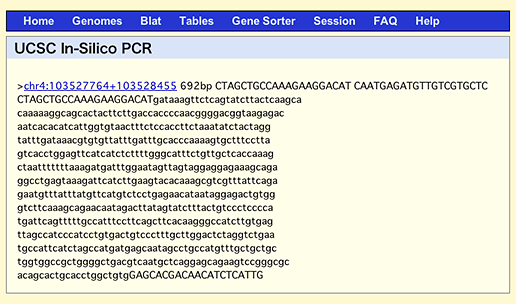
By the way, if you do the same search in UCSC In-Silico PCR, which is a famous and conventional web service to search for fragments amplified by the entered primers, it’ll come up with only one result with the size of 692 bp.
This discrepancy is attributable to the fact that UCSC service searches against genome, while GGRNA searches against transcripts. The above NFκB primers are designed to sandwich a 452 bp intron, so when you use genome DNA as a template, the product will be 240 + 452 = 692 (bp). When designing primers, sandwiching an intron is a good way: since the products derived from cDNA template and genomic DNA template will give different lengths, you can detect genomic DNA contamination in your template by simply looking at the product’s uniformity.
Search for nucleotide sequences included in figures
GGRNA works great for searching for nucleotide sequences appearing in figures of articles.
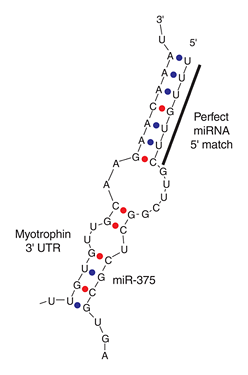
[Rajewsky et al. (2006) microRNA target predictions in animals. Nature Genetics 38, S8 – S13]
Left RNA strand shows the partial 3′ UTR sequence of myotrophin, which is the target of mice miR-375. Try searching with its partial sequence using GGRNA by selecting Mus musculus (mouse) and entering [ GUUGCAAGA ] to the search box (→ GGRNA). But it returns 322 hits, which is too much. So narrow them down by entering longer fragment [ GUUGCAAGAACAAA ] (→ GGRNA), and you’ll get 1 hit. Be aware that GGRNA treats U and T identically.
The sequence matched at position 3763, and CDS region is 279 – 635, meaning that miR-375 matches to farther downstream in 3′ UTR.
FYI, target sequence of miR-375 shown on the right side in the figure can be retrieved by entering about 13 letters, [ UUUGUUCGUUCGG ] (→ GGRNA).
Next.
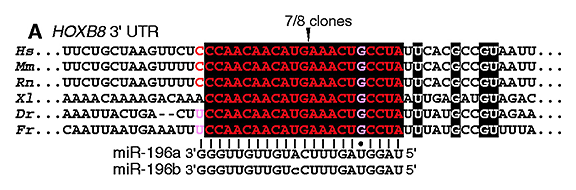
[Yekta et al. (2004) MicroRNA-directed cleavage of HOXB8 mRNA. Science 304, 594-596]
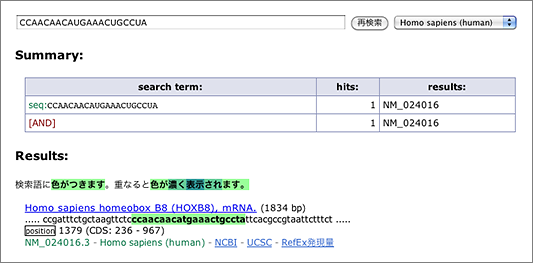
Let’s search using the letters in black background [ CCAACAACAUGAAACUGCCUA ] (in human, mouse, or rat as you like) (→ GGRNA) and it’ll return position 1379 of HOXB8 (NM_024016.3) (CDS: 236 – 967), confirming that they indeed match to 3′ UTR.
For your information, search operators such as comp: for searching for complementary sequences, both: for searching for both strands, seq1:, seq2:, and seq3: for the search allowing 1, 2, or 3 mismatches respectively, are available.
Search for siRNA off-target transcripts
In mammalian RNAi, 21 nt double-stranded short interfering RNA (siRNA) is used. However, if an siRNA sequence resembles to another unrelated gene, it may unexpectedly suppress that gene. This is referred to as an off-target effect. In siDirect (website for designing functional siRNA with reduced off-target effect, which I have launched), designed siRNA sequences (19-mer sequence on the guide strand counting from 5′ positions 2 to 20) are used for homology search and it returns a list of genes homologous to the sequences (with up to 3 mismatches). By the way, why I use the 19-mer rather than the full length (positions 1 to 21) is that when RNAi takes place, the nucleotide at the 5′ end of the guide strand is in the pocket of Mid-domain in Argonaute protein, and the nucleotide at the 3′ end binds to PAZ domain, making them irrelevant for target mRNA recognition. The number of mismatches enough to avoid off-target effect has not been determined yet: 1 mismatch is definitely not enough, and as the number of mismatches increase, the risk of off-target effect decreases. From the bioinformatics analysis, it is possible to design siRNAs that have at least 3 mismatches to any other unrelated genes than the target sequence (for approximately 10% of the entire gene), but it is hardly possible to design them with at least 4 mismatches.
Here we use the below siRNA sequence to search for homologous genes using GGRNA. This siRNA is designed to target a gene called claudin 17.
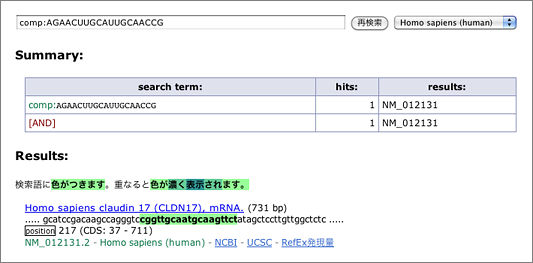
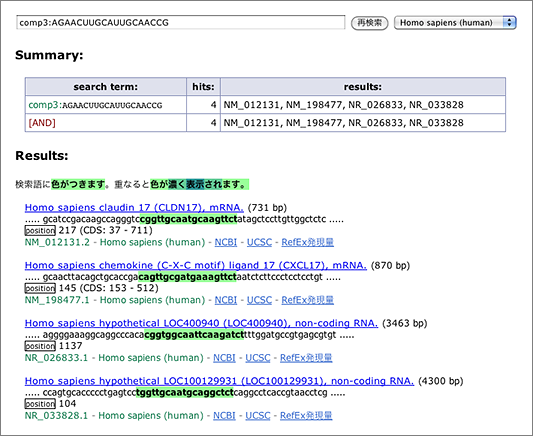
What we want to do here is to search for sequences that hybridize with 5′-AGAACUUGCAUUGCAACCG-3’, which is the 19-mer of the siRNA guide strand 5′-UAGAACUUGCAUUGCAACCGG-3′ (both ends removed), so let’s start with entering [ comp:AGAACUUGCAUUGCAACCG ] to the search box (→ GGRNA).
Claudin 17 (CLDN17; NM_012131.2), the target gene of this siRNA is the only result retrieved. Now let’s try again with allowing mismatches.
- allows up to 1 mismatch → [ comp1:AGAACUUGCAUUGCAACCG ] (→ GGRNA)
- allows up to 2 mismatches → [ comp2:AGAACUUGCAUUGCAACCG ] (→ GGRNA)
- allows up to 3 mismatches → [ comp3:AGAACUUGCAUUGCAACCG ] (→ GGRNA)
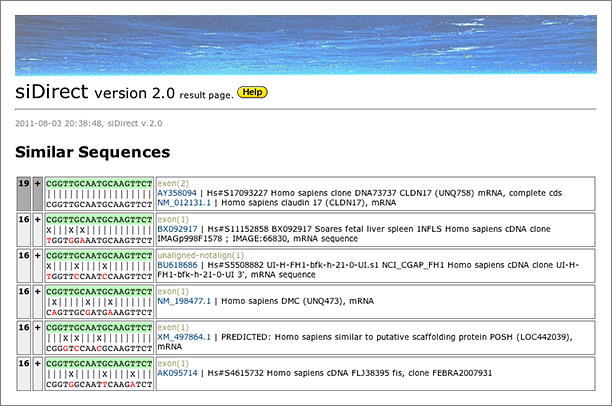
When we tolerated 3 mismatches, 3 other hits are finally retrieved. siDirect will return the results shown below, and the difference between GGRNA and siDirect comes from the versions of the sequence database used (GGRNA uses newer version of the database). The mismatched positions are better visualized in siDirect than in GGRNA: I’m planning to update GGRNA to display the mismatched nucleotides in different color like siDirect near future.
Of course, passenger strand can cause off-target effect as well as guide strand, so the passenger strand should also be searched. From the passenger strand 5′-GGUUGCAAUGCAAGUUCUAUA-3′, remove the nucleotides at both ends and search for sequences hybridizing with 5′-GUUGCAAUGCAAGUUCUAU-3′:
- perfect match → [ comp:GUUGCAAUGCAAGUUCUAU ] (→ GGRNA): no hit
- up to 1 mismatch → [ comp1:GUUGCAAUGCAAGUUCUAU ] (→ GGRNA): no hit
- up to 2 mismatches → [ comp2:GUUGCAAUGCAAGUUCUAU ] (→ GGRNA): no hit
- up to 3 mismatches → [ comp3:GUUGCAAUGCAAGUUCUAU ] (→ GGRNA): 5 hits
Here 3 mismatches had to be tolerated to return off-target candidates.
It is very rare to have few hits when you search 19-mer with 3 mismatches allowed. Thus the siRNA in the above example seems highly specific in terms of nucleotide sequence.
By the way, ‘seed’ sequences (7-mer nucleotides in positions 2 to 8 of guide strand) with higher Tm also have the risk of exerting off-target effect to genes whose 3′ UTR contain sequences identical to the seed. Please refer to the below paper or TogoTV guide for siDirect for further information. To design siRNAs with less off-target effect, you have to start with choosing sequences with lower seed Tm.
- Naito et al. (2009) siDirect 2.0: updated software for designing functional siRNA with reduced seed-dependent off-target effect. BMC Bioinformatics 10, 392 → full text
- TogoTV: Designing siRNA with siDirect: 2011 (in Japanese only)
Search for nucleotide sequences using microarray probe ID
As I have shown in the entry on 2/Jun/2011 “Nucleotide search using microarry probe ID” (in Japanese), when you enter microarray probe IDs, GGRNA will search for genes using the nucleotide sequences corresponding to the probe IDs. It also specifies the positions where probes hybridize.
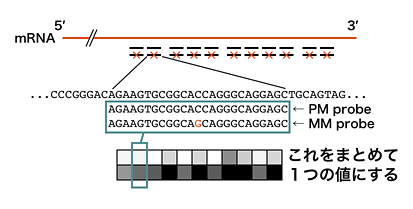
In particular, Affymetrix microarray uses eleven 25-mer perfect match (PM) probes, collectively called ‘probeset,’ to recognize single transcript. As shown below, Affymetrix also provides mismatch (MM) probes at the same positions as the 11 PM probes for background, but it is not used as often as before.
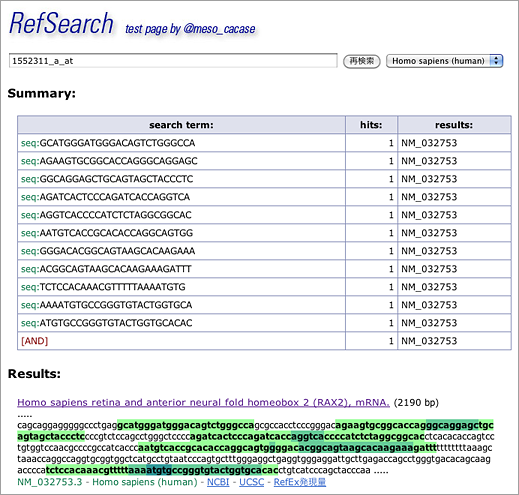
When probeset ID is entered in GGRNA like [ 1552311_a_at ] (→ GGRNA), GGRNA converts probeset ID into nucleotide sequences to perform search with:
[ GCATGGGATGGGACAGTCTGGGCCA ] +
[ AGAAGTGCGGCACCAGGGCAGGAGC ] +
[ GGCAGGAGCTGCAGTAGCTACCCTC ] +
[ AGATCACTCCCAGATCACCAGGTCA ] +
[ AGGTCACCCCATCTCTAGGCGGCAC ] +
[ AATGTCACCGCACACCAGGCAGTGG ] +
[ GGGACACGGCAGTAAGCACAAGAAA ] +
[ ACGGCAGTAAGCACAAGAAAGATTT ] +
[ TCTCCACAAACGTTTTTAAAATGTG ] +
[ AAAATGTGCCGGGTGTACTGGTGCA ] +
[ ATGTGCCGGGTGTACTGGTGCACAC ] .
The search returns RAX2 (NM_032753) gene. Click the title of the hit…
…and you will see that the 11 oligonucleotides hybridize with sequences close to 3′ end. Nucleotides with overlapping hits are shown in dark green.
Meanwhile, Agilent microarray uses one 60-mer probe to recognize single transcript. For example, searching with [ A_23_P101434 ] will return the following result (→ GGRNA):
Search for binding motifs of RNA-binding protein
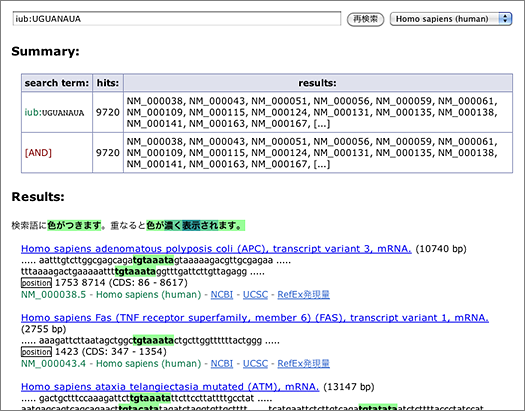
Degenerate motifs recoognized by RNA binding proteins can be searched using IUB codes (e.g., N, R, Y). For example, search for mRNA having PUM binding site UGUANAUA by entering [ iub:UGUANAUA ] to the search box (→ GGRNA: will take about 10 secs).
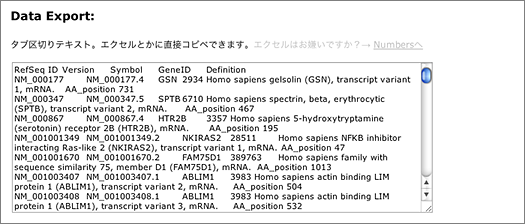
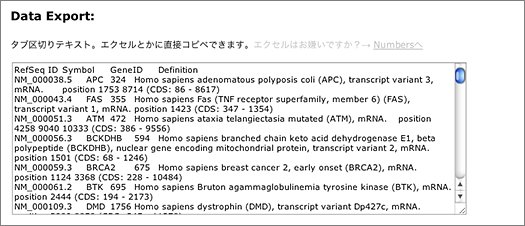
The search returns 9,720 hits. You can narrow them down by entering other keywords, or use the tab-delimited format provided at the bottom of the page for further analyses using other softwares.
- Comments (Close): 0
- Trackbacks: 0
Home > Archives > 2012-04
- Search
- Feeds
- Meta